Biological systems use complex macromolecular nanostructure networks to mediate a range of cellular functions, such as biomolecule synthesis, signal transduction, and gene expression and regulation, all with high efficiency and specificity. Many of these macromolecular systems have evolved through the spontaneous self-assembly of components into highly organized spatial structures. Mimicking these structures outside of the cell requires methods that offer nanoscale control over the organization of individual network components. We aim to apply the self-assembled molecular scaffolds to the organization of biomoleuclar networks with the intention of understanding fundamental mechanisms for biochemistry reactions as well as developing novel regulatory biocircuits. Research themes in our lab include:
Bionanotechnology:
DNA has recently emerged as an extremely promising material to organize molecules at the nanoscale. The reliability of base recognition, self-assembling behavior, and addressable surfaces of DNA nanostructures are of unparalleled value in systems of this size. DNA scaffolds have already been used to organize a variety of molecules, including nanoparticles and proteins. New bioconjugation chemistries make it possible to precisely position proteins and other biomolecules on DNA scaffolds, assembling biochemical reaction pathways with the ability to modulate inter-molecular interactions and the local environment.
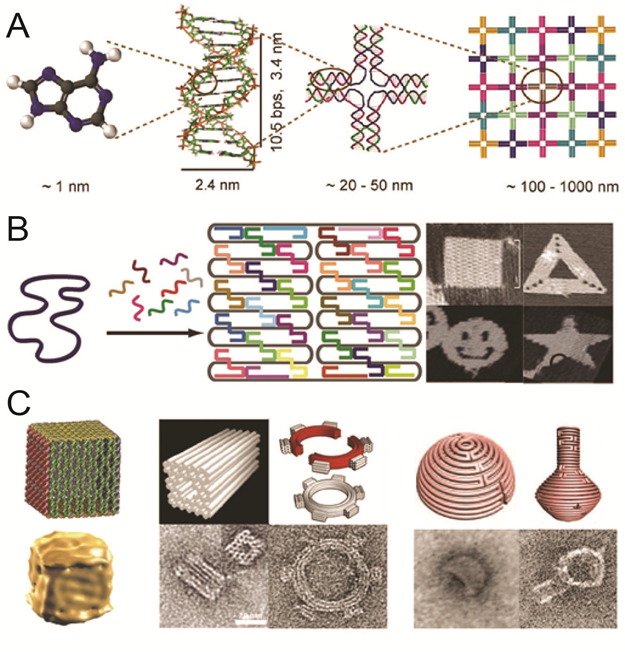
Organization the spatial arrangement of biomolecular network:
DNA-scaffold-directed assembly holds great promise to organize multi-enzyme systems with precise control of spatial patterning. Many of the enzymes in biochemical pathways are spatially organized to improve both reaction speed and specificity. Understanding the effect of spatial organization on enzymatic activity in multi-enzyme systems is not only fundamentally interesting, but also important for translating biochemical pathways to noncellular environments.
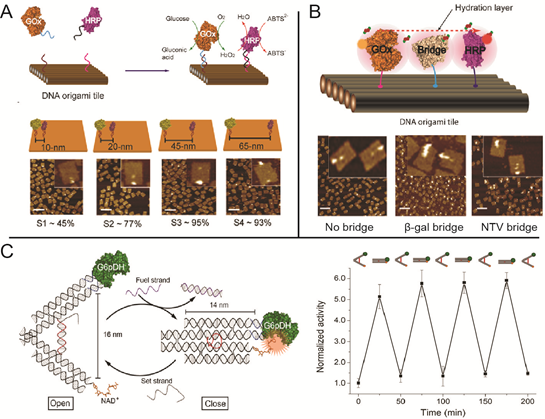
Molecular sensing circuits:
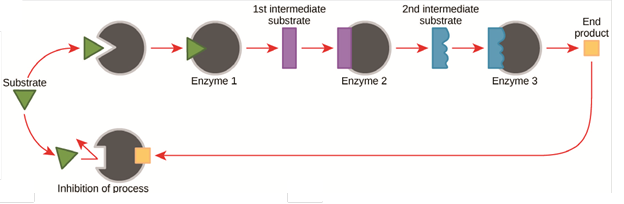
Cellular activities are directed by complex, multi-step pathways that exhibit extraordinary yield and specificity. The ability to regulate pathway activities in response to cellular environmental conditions (e.g. substrate levels, stimulants, etc.) plays a key role in metabolic functions, controlling the production of molecules and energy. Metabolic pathways have evolved feedback mechanisms to control the reactions, such as product feedback inhibition and allosteric modulation. The figure below shows a typical product feedback inhibition process for a multi-enzyme pathway in which the abundance of final product induces the inhibition of upstream enzymes to decrease the pathway rate. Some enzyme pathways also respond to important cofactor levels such as ATP or metal ions like magnesium and zinc.